Generating Tabular Synthetic Data Using GANs
Checkout mdedit.ai, AI powered Markdown Editor for tech writers
In this post, we will see how to generate tabular synthetic data using Generative adversarial networks(GANs). The goal is to generate synthetic data that is similar to the actual data in terms of statistics and demographics.
Introduction
It is important to ensure data privacy while publicly sharing information that contains sensitive information. There are numerous ways to tackle it and in this post we will use neural networks to generate synthetic data whose statistical features match the actual data.
We would be working with the Synthea dataset which is publicly available. Using the patients data from this dataset, we will try to generate synthetic data.
https://synthetichealth.github.io/synthea/
TL;DR
Checkout the Python Notebook if you are here just for the code.
https://colab.research.google.com/drive/1vBnSrTP8liPlnGg5ArzUk08oDLnERDbF?usp=sharing
Data Preprocessing
Firstly, download the publicly available synthea dataset and unzip it.
| !wget https://storage.googleapis.com/synthea-public/synthea_sample_data_csv_apr2020.zip | |
| !unzip synthea_sample_data_csv_apr2020.zip |
Remove unnecessary columns and encode all data
Next, read patients data and remove fields such as id, date, SSN, name etc. Note, that we are trying to generate synthetic data which can be used to train our deep learning models for some other tasks. For such a model, we don’t require fields like id, date, SSN etc.
| import pandas as pd | |
| df = pd.read_csv('csv/patients.csv') | |
| df.drop(['Id', 'BIRTHDATE', 'DEATHDATE', 'SSN', 'DRIVERS', 'PASSPORT', 'PREFIX', | |
| 'FIRST', 'ADDRESS', 'LAST', 'SUFFIX', 'MAIDEN','LAT', 'LON',], axis=1, inplace=True) | |
| print(df.columns) |
Index(['MARITAL', 'RACE', 'ETHNICITY', 'GENDER', 'BIRTHPLACE', 'CITY', 'STATE',
'COUNTY', 'ZIP', 'HEALTHCARE_EXPENSES', 'HEALTHCARE_COVERAGE'],
dtype='object')
Next, we will encode all categorical features to integer values. We are simply encoding the features to numerical values and are not using one hot encoding as its not required for GANs.
| df["MARITAL"] = df["MARITAL"].astype('category').cat.codes | |
| df["RACE"] = df["RACE"].astype('category').cat.codes | |
| df["ETHNICITY"] = df["ETHNICITY"].astype('category').cat.codes | |
| df["GENDER"] = df["GENDER"].astype('category').cat.codes | |
| df["BIRTHPLACE"] = df["BIRTHPLACE"].astype('category').cat.codes | |
| df["CITY"] = df["CITY"].astype('category').cat.codes | |
| df["STATE"] = df["STATE"].astype('category').cat.codes | |
| df["COUNTY"] = df["COUNTY"].astype('category').cat.codes | |
| df["ZIP"] = df["ZIP"].astype('category').cat.codes | |
| df.head() |
| MARITAL | RACE | ETHNICITY | GENDER | BIRTHPLACE | CITY | STATE | COUNTY | ZIP | HEALTHCARE_EXPENSES | HEALTHCARE_COVERAGE | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 4 | 0 | 1 | 136 | 42 | 0 | 6 | 2 | 271227.08 | 1334.88 |
| 1 | 0 | 4 | 1 | 1 | 61 | 186 | 0 | 8 | 132 | 793946.01 | 3204.49 |
| 2 | 0 | 4 | 1 | 1 | 236 | 42 | 0 | 6 | 3 | 574111.90 | 2606.40 |
| 3 | 0 | 4 | 1 | 0 | 291 | 110 | 0 | 8 | 68 | 935630.30 | 8756.19 |
| 4 | -1 | 4 | 1 | 1 | 189 | 24 | 0 | 12 | 125 | 598763.07 | 3772.20 |
Next, we will encode all
continious features to equally sized bins. First, lets find the minimum and maximum values for HEALTHCARE_EXPENSES and HEALTHCARE_COVERAGE and then create bins based on these values.
| HEALTHCARE_EXPENSES_MIN = df["HEALTHCARE_EXPENSES"].min() | |
| HEALTHCARE_EXPENSES_MAX = df["HEALTHCARE_EXPENSES"].max() | |
| print('Min and max healthcare expense', HEALTHCARE_EXPENSES_MIN, HEALTHCARE_EXPENSES_MAX) | |
| HEALTHCARE_COVERAGE_MIN = df["HEALTHCARE_COVERAGE"].min() | |
| HEALTHCARE_COVERAGE_MAX = df["HEALTHCARE_COVERAGE"].max() | |
| print('Min and max healthcare coverage', HEALTHCARE_COVERAGE_MIN, HEALTHCARE_COVERAGE_MAX) | |
Min and max healthcare expense 1822.1600000000005 2145924.400000002
Min and max healthcare coverage 0.0 927873.5300000022
Now, we encode HEALTHCARE_EXPENSES and HEALTHCARE_COVERAGE into bins using the pd.cut method. We use numpy’s linspace method to create equally sized bins.
| import numpy as np | |
| df_healthcare_expenses = pd.cut(df['HEALTHCARE_EXPENSES'], bins=np.linspace(HEALTHCARE_EXPENSES_MIN, HEALTHCARE_EXPENSES_MAX, 21), labels=False) | |
| df_healthcare_coverage = pd.cut(df['HEALTHCARE_COVERAGE'], bins=np.linspace(HEALTHCARE_COVERAGE_MIN, HEALTHCARE_COVERAGE_MAX, 21), labels=False) | |
| df.drop(["HEALTHCARE_EXPENSES", "HEALTHCARE_COVERAGE"], axis=1, inplace=True) | |
| df = pd.concat([df, df_healthcare_expenses, df_healthcare_coverage], axis=1) |
Transform the data
Next, we apply PowerTransformer on all the fields to get a Gaussian distribution for the data.
| from sklearn.preprocessing import PowerTransformer | |
| df[df.columns] = PowerTransformer(method='yeo-johnson', standardize=True, copy=True).fit_transform(df[df.columns]) | |
| print(df) |
MARITAL RACE ... HEALTHCARE_EXPENSES HEALTHCARE_COVERAGE
0 0.334507 0.461541 ... -0.819522 -0.187952
1 0.334507 0.461541 ... 0.259373 -0.187952
2 0.334507 0.461541 ... -0.111865 -0.187952
3 0.334507 0.461541 ... 0.426979 -0.187952
4 -1.275676 0.461541 ... -0.111865 -0.187952
... ... ... ... ... ...
1166 0.334507 -2.207146 ... 1.398831 -0.187952
1167 1.773476 0.461541 ... 0.585251 -0.187952
1168 1.773476 0.461541 ... 1.275817 5.320497
1169 0.334507 0.461541 ... 1.016430 -0.187952
1170 0.334507 0.461541 ... 1.275817 -0.187952
[1171 rows x 11 columns]
/usr/local/lib/python3.6/dist-packages/sklearn/preprocessing/_data.py:2982: RuntimeWarning: divide by zero encountered in log
loglike = -n_samples / 2 * np.log(x_trans.var())
Train the Model
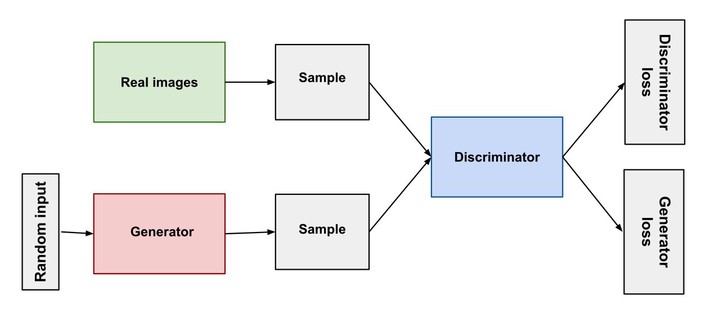
Next, lets define the neural network for generating synthetic data. We will be using a GAN network that comprises of an generator and discriminator that tries to beat each other and in the process learns the vector embedding for the data.
The model was taken from a Github repository where it is used to generate synthetic data on credit card fraud data.
| import os | |
| import numpy as np | |
| import tensorflow as tf | |
| from tensorflow.keras.layers import Input, Dense, Dropout | |
| from tensorflow.keras import Model | |
| from tensorflow.keras.optimizers import Adam | |
| class GAN(): | |
| def __init__(self, gan_args): | |
| [self.batch_size, lr, self.noise_dim, | |
| self.data_dim, layers_dim] = gan_args | |
| self.generator = Generator(self.batch_size).\ | |
| build_model(input_shape=(self.noise_dim,), dim=layers_dim, data_dim=self.data_dim) | |
| self.discriminator = Discriminator(self.batch_size).\ | |
| build_model(input_shape=(self.data_dim,), dim=layers_dim) | |
| optimizer = Adam(lr, 0.5) | |
| # Build and compile the discriminator | |
| self.discriminator.compile(loss='binary_crossentropy', | |
| optimizer=optimizer, | |
| metrics=['accuracy']) | |
| # The generator takes noise as input and generates imgs | |
| z = Input(shape=(self.noise_dim,)) | |
| record = self.generator(z) | |
| # For the combined model we will only train the generator | |
| self.discriminator.trainable = False | |
| # The discriminator takes generated images as input and determines validity | |
| validity = self.discriminator(record) | |
| # The combined model (stacked generator and discriminator) | |
| # Trains the generator to fool the discriminator | |
| self.combined = Model(z, validity) | |
| self.combined.compile(loss='binary_crossentropy', optimizer=optimizer) | |
| def get_data_batch(self, train, batch_size, seed=0): | |
| # # random sampling - some samples will have excessively low or high sampling, but easy to implement | |
| # np.random.seed(seed) | |
| # x = train.loc[ np.random.choice(train.index, batch_size) ].values | |
| # iterate through shuffled indices, so every sample gets covered evenly | |
| start_i = (batch_size * seed) % len(train) | |
| stop_i = start_i + batch_size | |
| shuffle_seed = (batch_size * seed) // len(train) | |
| np.random.seed(shuffle_seed) | |
| train_ix = np.random.choice(list(train.index), replace=False, size=len(train)) # wasteful to shuffle every time | |
| train_ix = list(train_ix) + list(train_ix) # duplicate to cover ranges past the end of the set | |
| x = train.loc[train_ix[start_i: stop_i]].values | |
| return np.reshape(x, (batch_size, -1)) | |
| def train(self, data, train_arguments): | |
| [cache_prefix, epochs, sample_interval] = train_arguments | |
| data_cols = data.columns | |
| # Adversarial ground truths | |
| valid = np.ones((self.batch_size, 1)) | |
| fake = np.zeros((self.batch_size, 1)) | |
| for epoch in range(epochs): | |
| # --------------------- | |
| # Train Discriminator | |
| # --------------------- | |
| batch_data = self.get_data_batch(data, self.batch_size) | |
| noise = tf.random.normal((self.batch_size, self.noise_dim)) | |
| # Generate a batch of new images | |
| gen_data = self.generator.predict(noise) | |
| # Train the discriminator | |
| d_loss_real = self.discriminator.train_on_batch(batch_data, valid) | |
| d_loss_fake = self.discriminator.train_on_batch(gen_data, fake) | |
| d_loss = 0.5 * np.add(d_loss_real, d_loss_fake) | |
| # --------------------- | |
| # Train Generator | |
| # --------------------- | |
| noise = tf.random.normal((self.batch_size, self.noise_dim)) | |
| # Train the generator (to have the discriminator label samples as valid) | |
| g_loss = self.combined.train_on_batch(noise, valid) | |
| # Plot the progress | |
| print("%d [D loss: %f, acc.: %.2f%%] [G loss: %f]" % (epoch, d_loss[0], 100 * d_loss[1], g_loss)) | |
| # If at save interval => save generated events | |
| if epoch % sample_interval == 0: | |
| #Test here data generation step | |
| # save model checkpoints | |
| model_checkpoint_base_name = 'model/' + cache_prefix + '_{}_model_weights_step_{}.h5' | |
| self.generator.save_weights(model_checkpoint_base_name.format('generator', epoch)) | |
| self.discriminator.save_weights(model_checkpoint_base_name.format('discriminator', epoch)) | |
| #Here is generating the data | |
| z = tf.random.normal((432, self.noise_dim)) | |
| gen_data = self.generator(z) | |
| print('generated_data') | |
| def save(self, path, name): | |
| assert os.path.isdir(path) == True, \ | |
| "Please provide a valid path. Path must be a directory." | |
| model_path = os.path.join(path, name) | |
| self.generator.save_weights(model_path) # Load the generator | |
| return | |
| def load(self, path): | |
| assert os.path.isdir(path) == True, \ | |
| "Please provide a valid path. Path must be a directory." | |
| self.generator = Generator(self.batch_size) | |
| self.generator = self.generator.load_weights(path) | |
| return self.generator | |
| class Generator(): | |
| def __init__(self, batch_size): | |
| self.batch_size=batch_size | |
| def build_model(self, input_shape, dim, data_dim): | |
| input= Input(shape=input_shape, batch_size=self.batch_size) | |
| x = Dense(dim, activation='relu')(input) | |
| x = Dense(dim * 2, activation='relu')(x) | |
| x = Dense(dim * 4, activation='relu')(x) | |
| x = Dense(data_dim)(x) | |
| return Model(inputs=input, outputs=x) | |
| class Discriminator(): | |
| def __init__(self,batch_size): | |
| self.batch_size=batch_size | |
| def build_model(self, input_shape, dim): | |
| input = Input(shape=input_shape, batch_size=self.batch_size) | |
| x = Dense(dim * 4, activation='relu')(input) | |
| x = Dropout(0.1)(x) | |
| x = Dense(dim * 2, activation='relu')(x) | |
| x = Dropout(0.1)(x) | |
| x = Dense(dim, activation='relu')(x) | |
| x = Dense(1, activation='sigmoid')(x) | |
| return Model(inputs=input, outputs=x) |
Next, lets define the training parameters for the GAN network. We would be using a batch size of 32 and train it for 5000 epochs.
| data_cols = df.columns |
| #Define the GAN and training parameters | |
| noise_dim = 32 | |
| dim = 128 | |
| batch_size = 32 | |
| log_step = 100 | |
| epochs = 5000+1 | |
| learning_rate = 5e-4 | |
| models_dir = 'model' | |
| df[data_cols] = df[data_cols] | |
| print(df.shape[1]) | |
| gan_args = [batch_size, learning_rate, noise_dim, df.shape[1], dim] | |
| train_args = ['', epochs, log_step] |
11
| !mkdir model |
mkdir: cannot create directory ‘model’: File exists
Finally, let’s run the training and see if the model is able to learn something.
| model = GAN | |
| #Training the GAN model chosen: Vanilla GAN, CGAN, DCGAN, etc. | |
| synthesizer = model(gan_args) | |
| synthesizer.train(df, train_args) |
[1;30;43mStreaming output truncated to the last 5000 lines.[0m
generated_data
101 [D loss: 0.324169, acc.: 85.94%] [G loss: 2.549267]
.
.
.
4993 [D loss: 0.150710, acc.: 95.31%] [G loss: 2.865143]
4994 [D loss: 0.159454, acc.: 95.31%] [G loss: 2.886763]
4995 [D loss: 0.159046, acc.: 95.31%] [G loss: 2.640226]
4996 [D loss: 0.150796, acc.: 95.31%] [G loss: 2.868319]
4997 [D loss: 0.170520, acc.: 95.31%] [G loss: 2.697939]
4998 [D loss: 0.161605, acc.: 95.31%] [G loss: 2.601780]
4999 [D loss: 0.156147, acc.: 95.31%] [G loss: 2.719781]
5000 [D loss: 0.164568, acc.: 95.31%] [G loss: 2.826339]
WARNING:tensorflow:Model was constructed with shape (32, 32) for input Tensor("input_1:0", shape=(32, 32), dtype=float32), but it was called on an input with incompatible shape (432, 32).
generated_data
After, 5000 epochs the models shows a training accuracy of 95.31% which sounds quite impressive.
| !mkdir model/gan | |
| !mkdir model/gan/saved |
mkdir: cannot create directory ‘model/gan’: File exists
mkdir: cannot create directory ‘model/gan/saved’: File exists
| #You can easily save the trained generator and loaded it aftwerwards | |
| synthesizer.save('model/gan/saved', 'generator_patients') |
Let’s take a look at the Generator and Discriminator models.
| synthesizer.generator.summary() |
Model: "model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_1 (InputLayer) [(32, 32)] 0
_________________________________________________________________
dense (Dense) (32, 128) 4224
_________________________________________________________________
dense_1 (Dense) (32, 256) 33024
_________________________________________________________________
dense_2 (Dense) (32, 512) 131584
_________________________________________________________________
dense_3 (Dense) (32, 11) 5643
=================================================================
Total params: 174,475
Trainable params: 174,475
Non-trainable params: 0
_________________________________________________________________
| synthesizer.discriminator.summary() |
Model: "model_1"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
input_2 (InputLayer) [(32, 11)] 0
_________________________________________________________________
dense_4 (Dense) (32, 512) 6144
_________________________________________________________________
dropout (Dropout) (32, 512) 0
_________________________________________________________________
dense_5 (Dense) (32, 256) 131328
_________________________________________________________________
dropout_1 (Dropout) (32, 256) 0
_________________________________________________________________
dense_6 (Dense) (32, 128) 32896
_________________________________________________________________
dense_7 (Dense) (32, 1) 129
=================================================================
Total params: 170,497
Trainable params: 0
Non-trainable params: 170,497
_________________________________________________________________
Evaluation
Now, that we have trained the model let’s see if the generated data is similar to the actual data.
We plot the generated data for some of the model steps and see how the plot for the generated data changes as the networks learns the embedding more accurately.
| models = {'GAN': ['GAN', False, synthesizer.generator]} |
| import matplotlib.pyplot as plt | |
| # Setup parameters visualization parameters | |
| seed = 17 | |
| test_size = 492 # number of fraud cases | |
| noise_dim = 32 | |
| np.random.seed(seed) | |
| z = np.random.normal(size=(test_size, noise_dim)) | |
| real = synthesizer.get_data_batch(train=df, batch_size=test_size, seed=seed) | |
| real_samples = pd.DataFrame(real, columns=data_cols) | |
| model_names = ['GAN'] | |
| colors = ['deepskyblue','blue'] | |
| markers = ['o','^'] | |
| col1, col2 = 'CITY', 'ETHNICITY' | |
| base_dir = 'model/' | |
| #Actual fraud data visualization | |
| model_steps = [ 0, 100, 200, 300, 400, 500, 1000, 2000, 3000, 4000, 5000] | |
| rows = len(model_steps) | |
| columns = 5 | |
| axarr = [[]]*len(model_steps) | |
| fig = plt.figure(figsize=(14,rows*3)) | |
| for model_step_ix, model_step in enumerate(model_steps): | |
| axarr[model_step_ix] = plt.subplot(rows, columns, model_step_ix*columns + 1) | |
| for group, color, marker in zip(real_samples.groupby('RACE'), colors, markers): | |
| plt.scatter( group[1][[col1]], group[1][[col2]], marker=marker, edgecolors=color, facecolors='none' ) | |
| plt.title('Actual Patients Data') | |
| plt.ylabel(col2) # Only add y label to left plot | |
| plt.xlabel(col1) | |
| xlims, ylims = axarr[model_step_ix].get_xlim(), axarr[model_step_ix].get_ylim() | |
| if model_step_ix == 0: | |
| legend = plt.legend() | |
| legend.get_frame().set_facecolor('white') | |
| i=0 | |
| [model_name, with_class, generator_model] = models['GAN'] | |
| generator_model.load_weights( base_dir + '_generator_model_weights_step_'+str(model_step)+'.h5') | |
| ax = plt.subplot(rows, columns, model_step_ix*columns + 1 + (i+1) ) | |
| g_z = generator_model.predict(z) | |
| gen_samples = pd.DataFrame(g_z, columns=data_cols) | |
| gen_samples.to_csv('Generated_sample.csv') | |
| plt.scatter( gen_samples[[col1]], gen_samples[[col2]], marker=markers[0], edgecolors=colors[0], facecolors='none' ) | |
| plt.title("Generated Data") | |
| plt.xlabel(data_cols[0]) | |
| ax.set_xlim(xlims), ax.set_ylim(ylims) | |
| plt.suptitle('Comparison of GAN outputs', size=16, fontweight='bold') | |
| plt.tight_layout(rect=[0.075,0,1,0.95]) | |
| # Adding text labels for traning steps | |
| vpositions = np.array([ i._position.bounds[1] for i in axarr ]) | |
| vpositions += ((vpositions[0] - vpositions[1]) * 0.35 ) | |
| for model_step_ix, model_step in enumerate( model_steps ): | |
| fig.text( 0.05, vpositions[model_step_ix], 'training\nstep\n'+str(model_step), ha='center', va='center', size=12) | |
| plt.savefig('Comparison_of_GAN_outputs.png') |
No handles with labels found to put in legend.
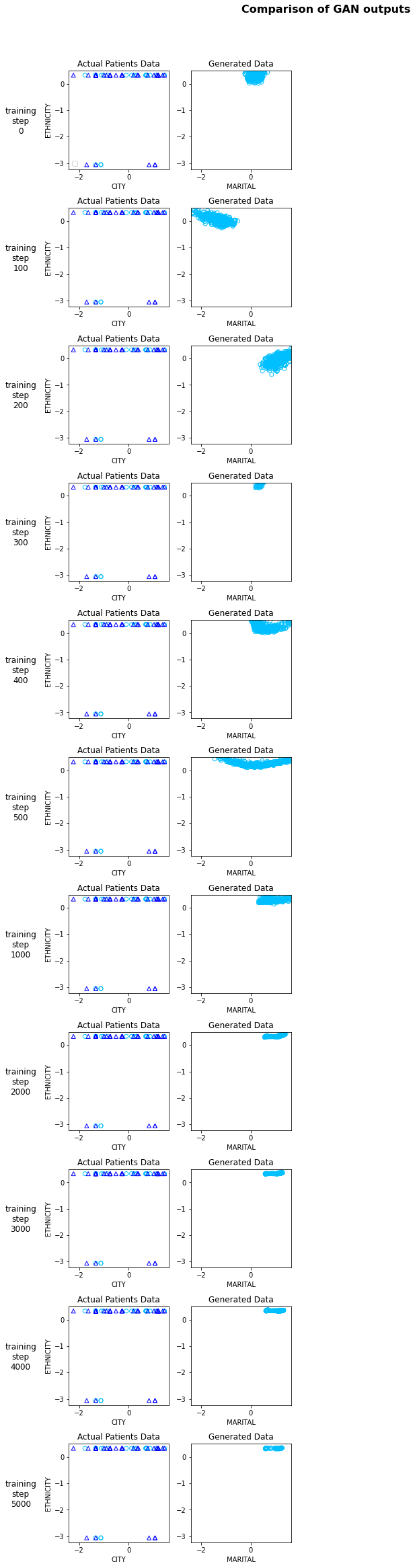
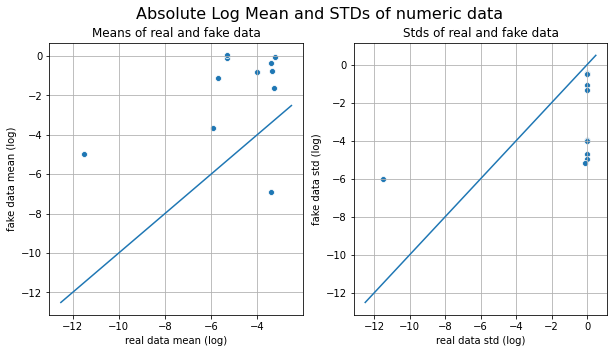
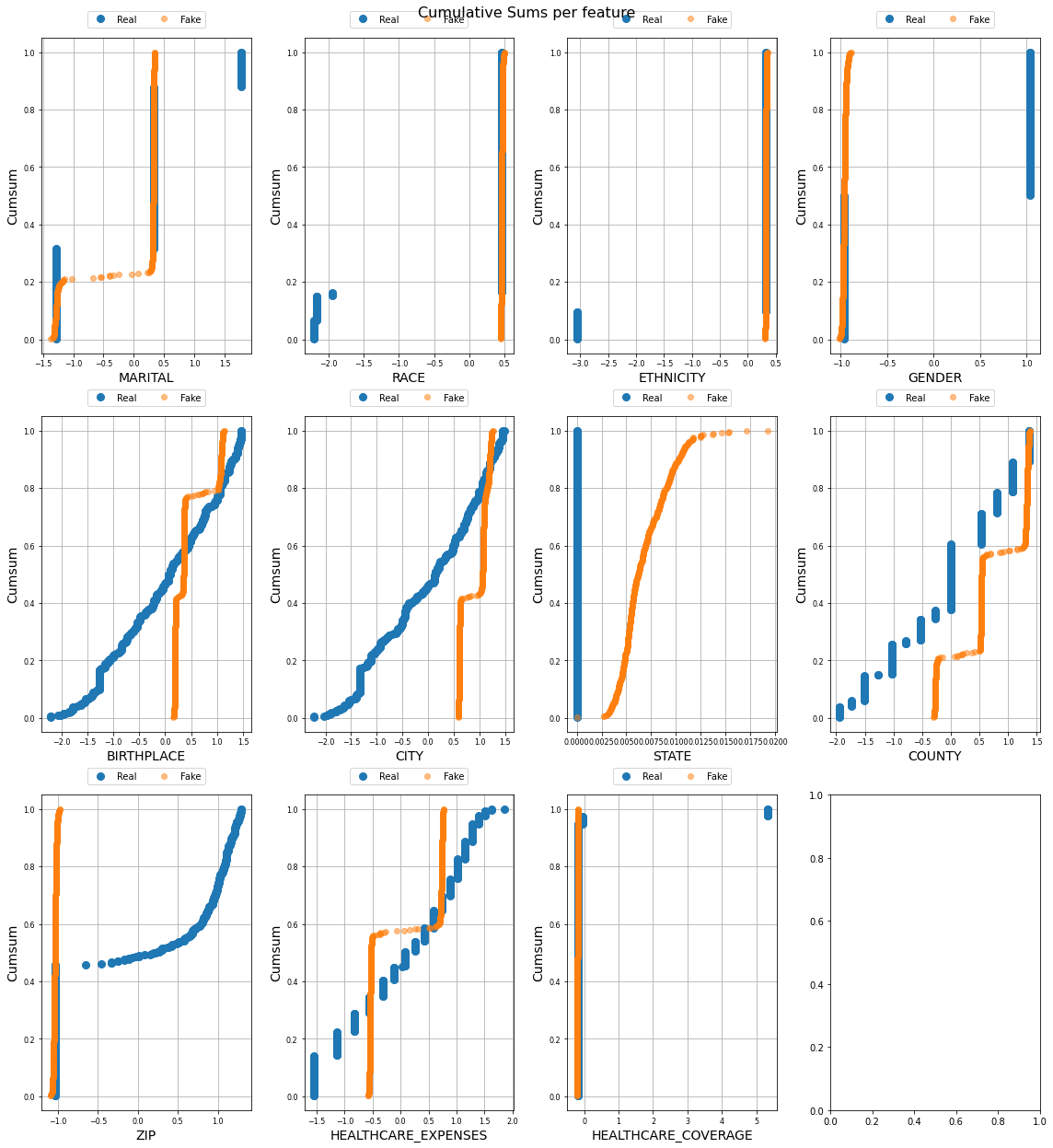
Now let’s try to do a feature by feature comparision between the generated data and the actual data. We will use python’s table_evaluator library to compare the features.
| !pip install table_evaluator | |
| gen_df.drop('Unnamed: 0', axis=1, inplace=True) | |
| print(gen_df.columns) | |
| print(df.shape, gen_df.shape) |
Index(['MARITAL', 'RACE', 'ETHNICITY', 'GENDER', 'BIRTHPLACE', 'CITY', 'STATE',
'COUNTY', 'ZIP', 'HEALTHCARE_EXPENSES', 'HEALTHCARE_COVERAGE'],
dtype='object')
(1171, 11) (492, 11)
We call the visual_evaluation method to compare the actual date(df) and the generated data(gen_df).
| from table_evaluator import load_data, TableEvaluator | |
| cat_cols = ['MARITAL', 'RACE', 'ETHNICITY', 'GENDER', 'BIRTHPLACE', 'CITY', 'STATE', 'COUNTY', 'ZIP'] | |
| print(len(df), len(gen_df)) | |
| table_evaluator = TableEvaluator(df, gen_df) | |
| table_evaluator.visual_evaluation() |
1171 492
/usr/local/lib/python3.6/dist-packages/seaborn/distributions.py:283: UserWarning: Data must have variance to compute a kernel density estimate.
warnings.warn(msg, UserWarning)
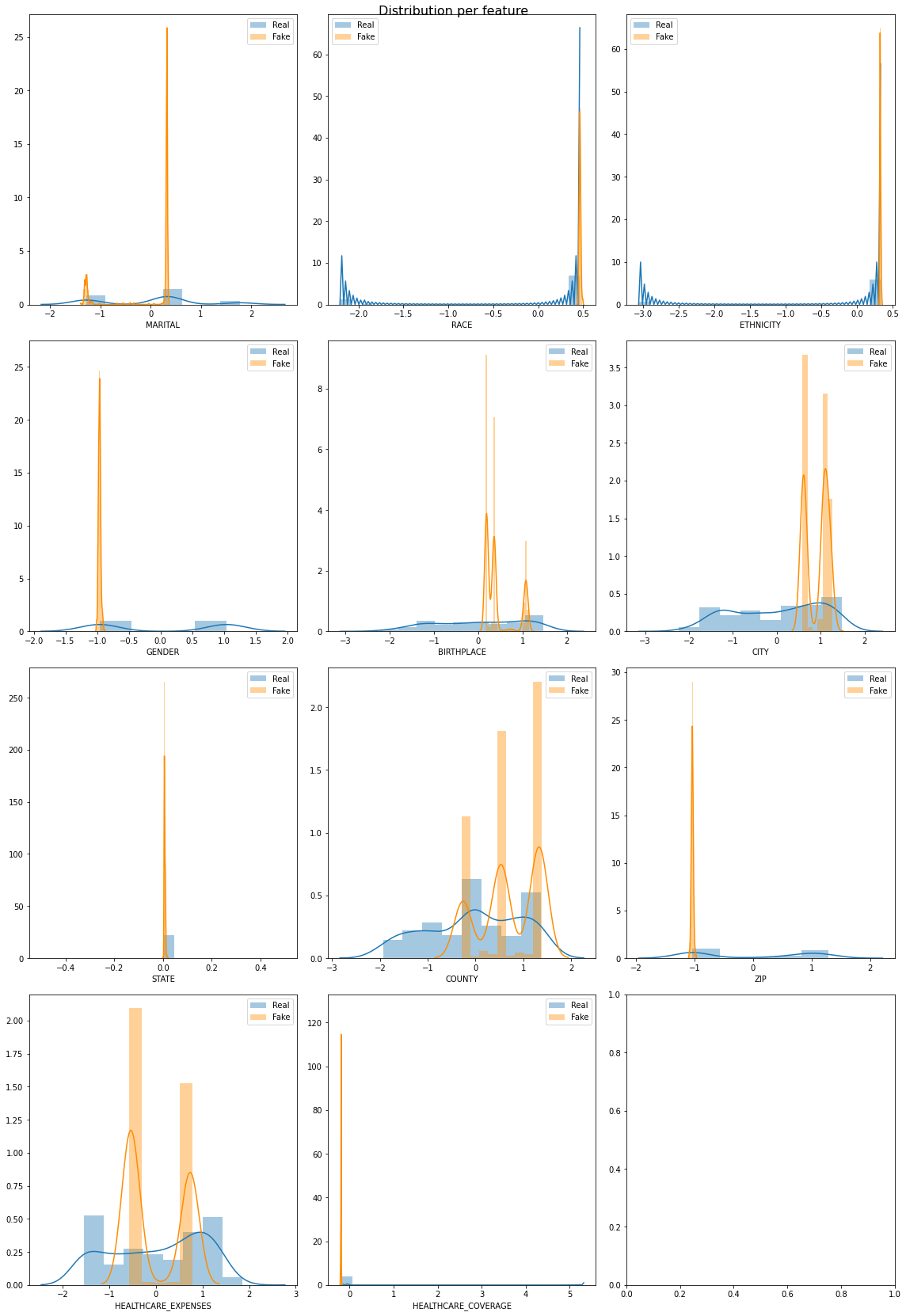

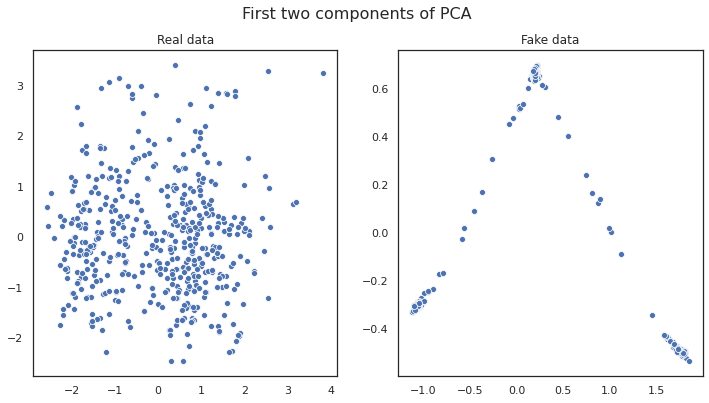
Conclusion
Some of the features in the syntehtic data match closely with actual data but there are some other features which were not learnt perfectly by the model. We can keep playing with the model and its hyperparameters to improve the model further.
This post demonstrates that its fairly simply to use GANs to generate synthetic data where the actual data is sensitive in nature and can’t be shared publicly.